Archive detail
Refined model for reliable prediction of invasion dynamics
January 16, 2014 |
In today’s globalized world, where people and goods move freely around the globe, animals, plants and microorganisms can also be transported almost anywhere within a short time and thus colonize new habitats thousands of kilometres away from their original territory. Control measures can only be implemented in the right place at the right time if the speed and patterns of such invasions can be reliably predicted.
Widely applied model
To predict the mean speed of invasion fronts, ecologists use a model originally developed in 1937 by the biologist R. A. Fisher and the mathematician A. N. Kolmogorov. Underlying this model are the processes of reaction (reproduction of organisms) and diffusion (movement of individuals in space); similar models are also applied in fields such as oncology (to describe tumour growth), as well as in physics and chemistry. Such modelling requires only two parameters, which can be readily determined: the diffusion coefficient and the rate of reproduction of the organisms in question.
Although the strengths of the ecological model are recognized, it has suffered from two decisive weakness: it has not previously been tested under controlled conditions, and it could not account for the variability in dispersal rates observed in nature. A team of researchers led by Eawag biologist Florian Altermatt and EPFL physicist Andrea Giometto have now added new elements to the model and performed laboratory experiments to test its predictions.
Dispersal in linear landscapes
Fluctuations in demographic processes are not attributable solely to environmental factors – under identical conditions, not every individual will produce the same number of offspring. Accordingly, the scientists modified the mathematical model by including a stochastic differential equation to account for random individual differences in reproduction within a population. With this modified model, variability in the speed of invasion fronts can also be predicted.
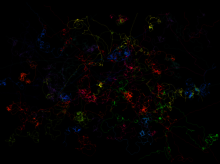
The modelling data were then compared with the results of microcosm experiments, in which a two-metre-long “linear landscape” (Plexiglas channel filled with nutrient medium) was used to study the dispersal of freshwater ciliates. The movements of individual organisms were recorded by video cameras (see figure), which made it possible to calculate the diffusion coefficient. The rate of dispersal towards the uncolonized end of the channel was determined by measuring cell density profiles.
The results of the laboratory experiments agree closely with the simulations. Thanks to these experiments, the scientists hope that – using the refined model – it will soon also be possible to make rapid, reliable predictions of colonization dynamics in natural environments with just a few simple measurements. Their findings were published January 7th, 2014 in the Proceedings of the National Academy of Sciences
Improving survival chances for threatened species
In contrast to efforts to control invasive species, conservation measures are designed to promote the recovery of locally extinct or severely reduced animal or plant populations. In 1988, US scientists used the Fisher-Kolmogorov model to study the Californian sea otter population along the Pacific coastline. In the early 1900s, as a result of hunting, the total number of sea otters had fallen to less than 100 at a single site. However, strict conservation regulations proved effective, and the otters began to spread north- and southwards again. The model yielded good predictions of the rate of range expansion and recolonization of former habitats. Giometto and his colleagues hope that this tool, now further refined, should also be able to predict variability in recolonization rates, allowing conservation measures to be optimized.
Local movement patterns of ciliates (Tetrahymena sp.) over a period of about a minute and a distance of a few millimetres. Each colour represents the trajectory of a different individual. By studying local movement and reproduction behaviour, it is possible to predict invasion dynamics over multiple generations.
A video microscope system (in the background) was used to record local movement patterns; Plexiglas channels (front) were used to determine the rate of dispersal of ciliates.
Further information
Florian Altermatt: +41 58 765 5592 (German and English); florian.altermatt@cluttereawag.ch
Andrea Giometto: +41 21 69 38027 (French, Italian and English); andrea.giometto@clutterepfl.ch http://echo.epfl.ch
Original publication
Giometto A, Rinaldo A, Carrara F & Altermatt F. Emerging predictable features of replicated biological invasion fronts. Proceedings of the National Academy of Sciences, DOI:10.1073/pnas.1321167110.
http://www.pnas.org/content/111/1/297.full.pdf+html