Department Environmental Toxicology
Development of protein marker panel to monitor chemical-induced molecular responses and damage progression in fish cells to inform animal-free prediction of chemical toxicity
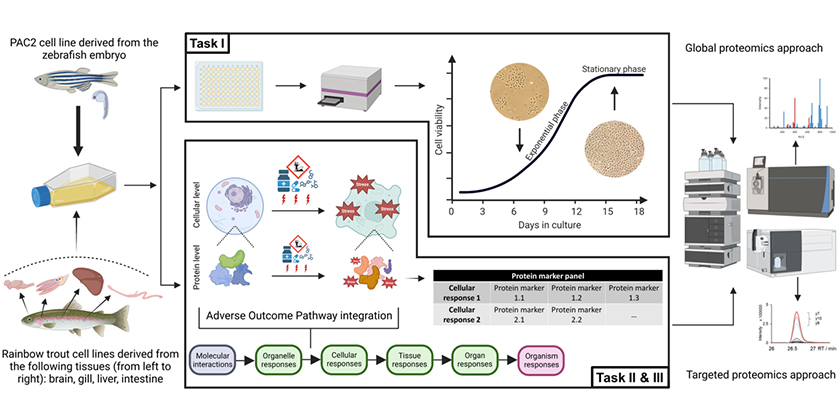
Environmental risk assessment often requires data on aquatic toxicity in fish. Currently, these data are still derived mostly with in vivo testing in juvenile or adult fish. These tests are resource intensive and use large numbers of animals, thus raising both economic and ethical concerns. Furthermore, as these tests rely on the apical endpoints such as mortality or growth, the obtained results provide little mechanistic information, which limits their usefulness for extrapolation to untested chemicals or across species. Alternative toxicity testing methods that use permanent fish cell lines (in vitro) as proxies for fish organism’s responses to chemical exposure in vivo offer the possibility to address some of these concerns, providing an approach to perform aquatic toxicity testing in an animal-free way. However, currently existing fish cell line-based tests still largely function as a “black box”, since they rely on a small set of molecular/cellular endpoints being measured and thus provide only a limited information on the mechanisms of toxicity.
This PhD project aims to expand the understanding of (i) functional capacities and tissue specificity of selected fish cell lines and (ii) their molecular responses initiated under chemical exposure, focusing specifically on the proteome level. It is part of a larger project that aims to develop a modular, computationally-linked framework that integrates different fish cell lines and mechanistic endpoints to allow fit-for-purpose animal-free prediction of chemical toxicity to fish, which is funded by the Swiss National Research Program 79 “Advancing 3Rs – Research, Animals and Society”.
To unravel the functional capacity and understand the degree of tissue specificity retained by permanent fish cell lines, mass spectrometry-based global proteomics analysis will be performed to analyse protein expression in the cells collected at the exponential and stationary growth stages of the cell culture cycle. Several fish cell lines of zebrafish and rainbow trout origin will be analysed. Proteomics and transcriptomics data (collected by another PhD student on this project) will then be integrated to understand the range of molecular components and pathways expressed by each cell line and perform a comparative analysis across the species- and tissue-origins.
To enable a detailed study of molecular responses induced by chemical exposure, a protein marker panel will be developed, which will include zebrafish proteins that are characteristic of certain toxicity mechanisms (such as e.g. oxidative stress) as well as toxicity-induced effects (e.g., energy metabolism changes or cytoskeleton rearrangements) occurring in the cells as the toxicity unfolds. Targeted proteomics assays will be established for candidate protein markers by means of selected reaction monitoring (SRM), which allows for higher sensitivity and faster sample throughput compared to global proteomics. Changes in the abundance of these proteins will then be measured in the zebrafish PAC2 cells in response to reference chemicals known to induce specific toxicity mechanisms, with the aim to obtain a detailed, concentration- and time-resolved understanding of the molecular changes that underlie cellular transition from adaptive to adverse responses, leading to the progression of toxicity within the chemically exposed cells. Upon successful establishment of the initial proof-of-principle protein marker panel, the investigations will expand to cover additional mechanisms and chemicals, as well as to comparative exploration of protein responses in rainbow trout cell lines. Obtained mechanistic and quantitative data will also be integrated into respective adverse outcome pathways.





